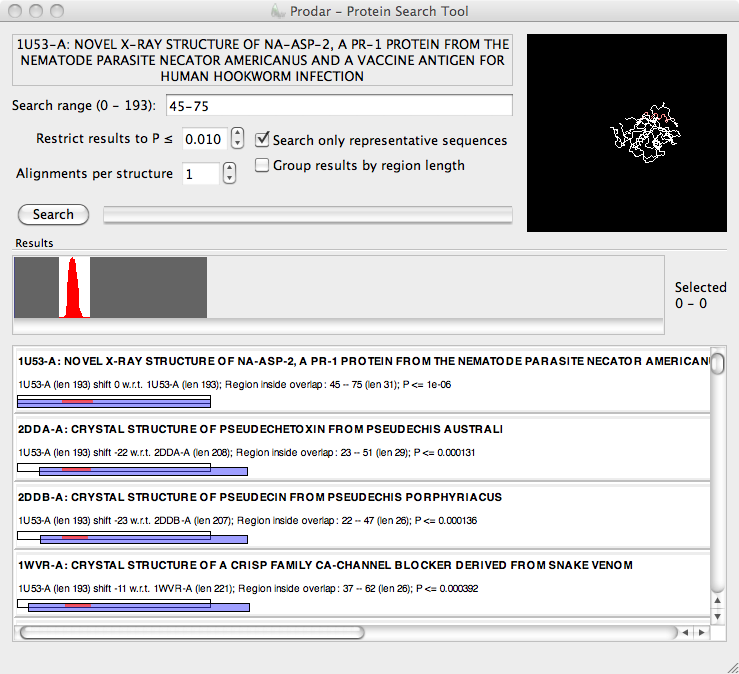

Prodar (Protein Radar) is a search application that queries the PDB for candidate structural alignments. The input to the search is a protein backbone structure read from a standard (text) PDB file, and the results returned are based solely on structural similarity of the backbone without any regard to sequence information. Searches are extremely fast, typically taking far less than a minute to complete. A highly-compressed and processed snapshot of the PDB is included with the download.
Prodar identifies partial matches, such that a relatively small section of the query structure may be matched against sections of other structures in the PDB regardless of the relative size of the chains. The intention is that the tool automatically identifies common domains and motifs in otherwise dissimilar protein structures. These can then be structurally aligned in a different tool to find an RMSD (Prodar just makes suggestions).
Prodar encourages an interactive style of discovery, where searches are rapidly refined and rerun. It is experimental, and I believe the signal processing techniques employed are novel in bioinformatics for this purpose.

Please use the Prodar Sourceforge Page, and consult the User Guide.
New snapshot of the PDB (Jan 2014).[Tue Feb 12 2013]
New snapshot of the PDB (Jan 2013).[Tue Apr 3 2012]
Prodar 1.5 is now released for Mac OS X. It includes a more recent snapshot of the PDB (March 2012), and a fix for PDB files exported from MOLMOL.[Mon Feb 27 2012]
Prodar 1.4 is now released for both Mac OS X and Windows.[Tue Jan 10 2012]
Prodar 1.3 is now released for both Mac OS X and Windows.